

| Field | Data |
|---|---|
| UniProt Accession: | O00459 |
| Gene Name: | PIK3R2 |
| Gene Synonim: | |
| GAP/GEF: | GAP |
| Description: | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase regulatory subunit beta) (PI3K regulatory subunit beta) (PtdIns-3-kinase regulatory subunit beta) (Phosphatidylinositol 3-kinase 85 kDa regulatory subunit beta) (PI3-kinase subunit p85-beta) (PtdIns-3-kinase regulatory subunit p85-beta) |
| Keywords: | 3D-structure;Complete proteome;Disease mutation;Phosphoprotein;Polymorphism;Protein transport;Reference proteome;Repeat;SH2 domain;SH3 domain;Stress response;Transport;Ubl conjugation |
| Entrez ID: | 5296 |
| Ensembl Gene ID: | ENSG00000105647 |
| Ensembl Protein ID: | ENSP00000477864 |
| Our Specificity | |
| Literature Specificity | None |
| Domain Composition: |
 |
| GO ID | Description |
|---|---|
| GO:0005634 | nucleus | GO:0005829 | cytosol | GO:0005942 | phosphatidylinositol 3-kinase complex |
| GO ID | Description |
|---|---|
| GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity | GO:0016303 | 1-phosphatidylinositol-3-kinase activity | GO:0030971 | receptor tyrosine kinase binding | GO:0005096 | GTPase activator activity | GO:0019903 | protein phosphatase binding | GO:0035014 | phosphatidylinositol 3-kinase regulator activity | GO:0005515 | protein binding | GO:0046982 | protein heterodimerization activity | GO:0001784 | phosphotyrosine residue binding |
| GO ID | Description |
|---|---|
| GO:0001678 | cellular glucose homeostasis | GO:0051056 | regulation of small GTPase mediated signal transduction | GO:0050852 | T cell receptor signaling pathway | GO:0050900 | leukocyte migration | GO:0007165 | signal transduction | GO:0032869 | cellular response to insulin stimulus | GO:0043409 | negative regulation of MAPK cascade | GO:0043547 | positive regulation of GTPase activity | GO:0046854 | phosphatidylinositol phosphorylation | GO:0006661 | phosphatidylinositol biosynthetic process | GO:0010506 | regulation of autophagy | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity | GO:0008286 | insulin receptor signaling pathway | GO:0038111 | interleukin-7-mediated signaling pathway | GO:0006810 | transport | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | GO:0014065 | phosphatidylinositol 3-kinase signaling | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | GO:0048015 | phosphatidylinositol-mediated signaling | GO:0042993 | positive regulation of transcription factor import into nucleus | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | GO:0038095 | Fc-epsilon receptor signaling pathway | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | GO:2001275 | positive regulation of glucose import in response to insulin stimulus | GO:0015031 | protein transport | GO:0034976 | response to endoplasmic reticulum stress |
| Reactome ID | Description |
|---|---|
| R-HSA-1660499 | Synthesis of PIPs at the plasma membrane | R-HSA-198203 | PI3K/AKT activation | R-HSA-4420097 | VEGFA-VEGFR2 Pathway | R-HSA-114604 | GPVI-mediated activation cascade | R-HSA-2219530 | Constitutive Signaling by Aberrant PI3K in Cancer | R-HSA-186763 | Downstream signal transduction | R-HSA-2730905 | Role of LAT2/NTAL/LAB on calcium mobilization | R-HSA-1266695 | Interleukin-7 signaling | R-HSA-1257604 | PIP3 activates AKT signaling | R-HSA-8853659 | RET signaling | R-HSA-389357 | CD28 dependent PI3K/Akt signaling | R-HSA-194840 | Rho GTPase cycle | R-HSA-388841 | Costimulation by the CD28 family | R-HSA-2029485 | Role of phospholipids in phagocytosis | R-HSA-202424 | Downstream TCR signaling | R-HSA-416482 | G alpha (12/13) signalling events | R-HSA-912526 | Interleukin receptor SHC signaling | R-HSA-1433557 | Signaling by SCF-KIT | R-HSA-392451 | G beta:gamma signalling through PI3Kgamma | R-HSA-2424491 | DAP12 signaling | R-HSA-512988 | Interleukin-3, 5 and GM-CSF signaling | R-HSA-912631 | Regulation of signaling by CBL | R-HSA-416476 | G alpha (q) signalling events | R-HSA-109704 | PI3K Cascade | R-HSA-210993 | Tie2 Signaling | R-HSA-6811558 | PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | R-HSA-373753 | Nephrin family interactions |
| Kegg ID | Description |
|---|---|
| hsa04152 | AMPK signaling pathway | hsa04550 | Signaling pathways regulating pluripotency of stem cells | hsa04611 | Platelet activation | hsa04510 | Focal adhesion | hsa04923 | Regulation of lipolysis in adipocytes | hsa05212 | Pancreatic cancer | hsa04650 | Natural killer cell mediated cytotoxicity | hsa05210 | Colorectal cancer | hsa05211 | Renal cell carcinoma | hsa05218 | Melanoma | hsa04024 | cAMP signaling pathway | hsa04630 | Jak-STAT signaling pathway | hsa05418 | Fluid shear stress and atherosclerosis | hsa04140 | Autophagy - animal | hsa04670 | Leukocyte transendothelial migration | hsa04725 | Cholinergic synapse | hsa05231 | Choline metabolism in cancer | hsa04068 | FoxO signaling pathway | hsa04380 | Osteoclast differentiation | hsa04066 | HIF-1 signaling pathway | hsa04926 | Relaxin signaling pathway | hsa04150 | mTOR signaling pathway | hsa04062 | Chemokine signaling pathway | hsa04960 | Aldosterone-regulated sodium reabsorption | hsa04370 | VEGF signaling pathway | hsa01521 | EGFR tyrosine kinase inhibitor resistance | hsa01522 | Endocrine resistance | hsa01524 | Platinum drug resistance | hsa04914 | Progesterone-mediated oocyte maturation | hsa05142 | Chagas disease (American trypanosomiasis) | hsa04218 | Cellular senescence | hsa05166 | HTLV-I infection | hsa05146 | Amoebiasis | hsa05169 | Epstein-Barr virus infection | hsa05213 | Endometrial cancer | hsa04211 | Longevity regulating pathway | hsa04210 | Apoptosis | hsa04213 | Longevity regulating pathway - multiple species | hsa04810 | Regulation of actin cytoskeleton | hsa04015 | Rap1 signaling pathway | hsa05161 | Hepatitis B | hsa05162 | Measles | hsa04662 | B cell receptor signaling pathway | hsa04917 | Prolactin signaling pathway | hsa04666 | Fc gamma R-mediated phagocytosis | hsa04072 | Phospholipase D signaling pathway | hsa05100 | Bacterial invasion of epithelial cells | hsa04973 | Carbohydrate digestion and absorption | hsa05205 | Proteoglycans in cancer | hsa04915 | Estrogen signaling pathway | hsa05206 | MicroRNAs in cancer | hsa04620 | Toll-like receptor signaling pathway | hsa05200 | Pathways in cancer | hsa05203 | Viral carcinogenesis | hsa04071 | Sphingolipid signaling pathway | hsa04070 | Phosphatidylinositol signaling system | hsa04910 | Insulin signaling pathway | hsa05224 | Breast cancer | hsa04750 | Inflammatory mediator regulation of TRP channels | hsa05222 | Small cell lung cancer | hsa05221 | Acute myeloid leukemia | hsa05220 | Chronic myeloid leukemia | hsa04668 | TNF signaling pathway | hsa04151 | PI3K-Akt signaling pathway | hsa04919 | Thyroid hormone signaling pathway | hsa04930 | Type II diabetes mellitus | hsa04931 | Insulin resistance | hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | hsa04933 | AGE-RAGE signaling pathway in diabetic complications | hsa04660 | T cell receptor signaling pathway | hsa05167 | Kaposi's sarcoma-associated herpesvirus infection | hsa05215 | Prostate cancer | hsa05226 | Gastric cancer | hsa05223 | Non-small cell lung cancer | hsa05160 | Hepatitis C | hsa05164 | Influenza A | hsa05225 | Hepatocellular carcinoma | hsa04014 | Ras signaling pathway | hsa04722 | Neurotrophin signaling pathway | hsa05165 | Human papillomavirus infection | hsa04360 | Axon guidance | hsa05230 | Central carbon metabolism in cancer | hsa05214 | Glioma | hsa04664 | Fc epsilon RI signaling pathway | hsa04012 | ErbB signaling pathway |
| Field | Data |
|---|---|
| Localization in this study: | focal_adhesions |
| Localization in the literature: |
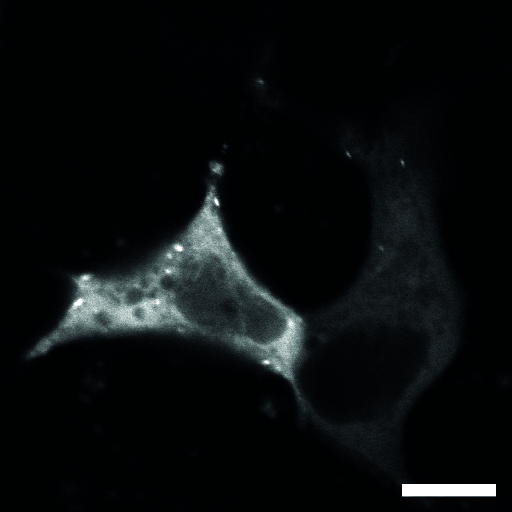
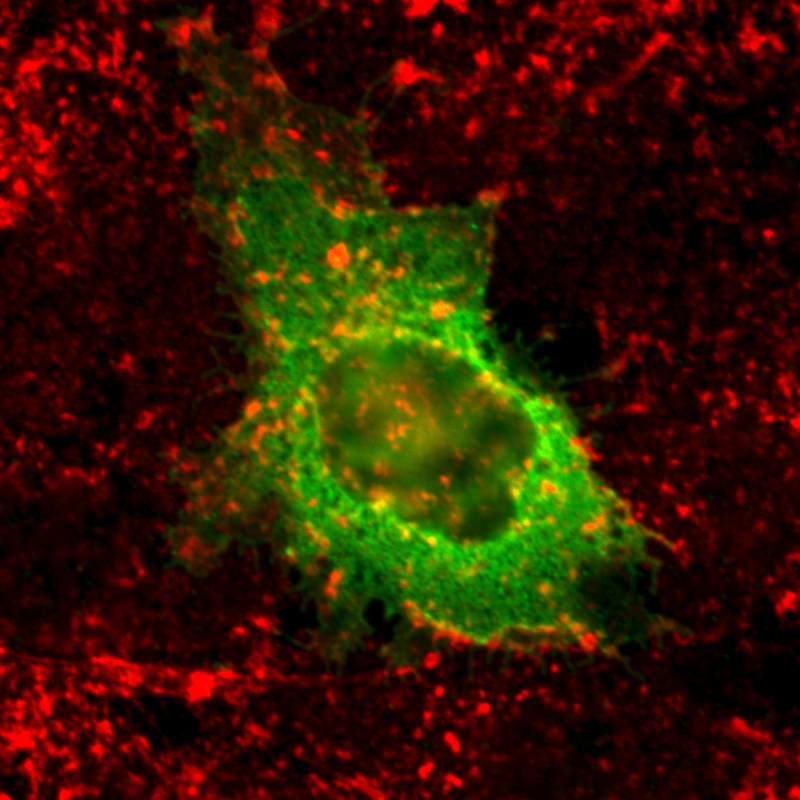
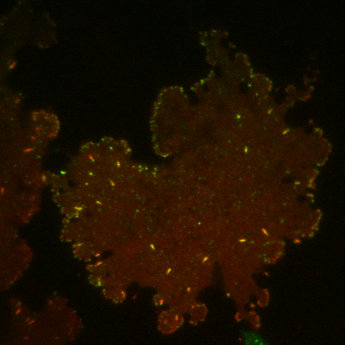
| organism | orgs |
|---|---|
| Organism: | Mus musculus |