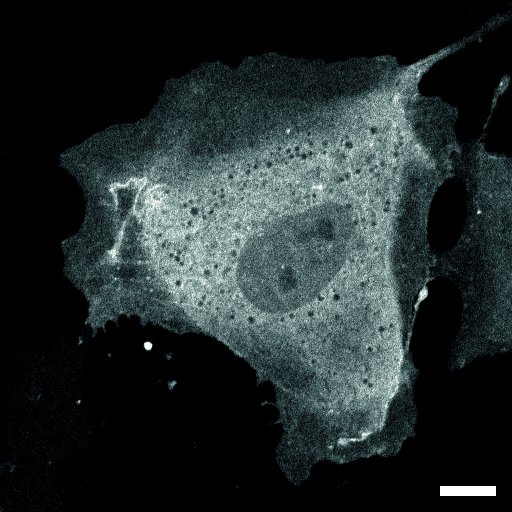
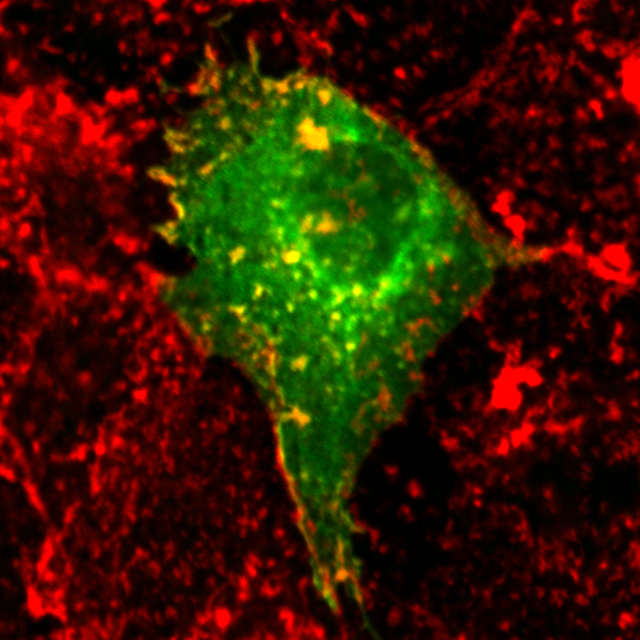
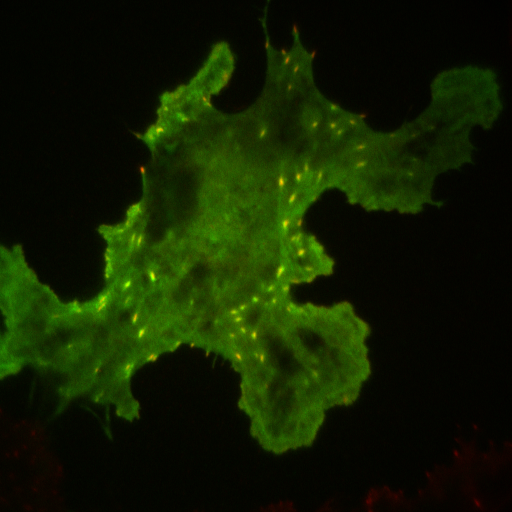
| organism |
orgs |
| Organism: |
Homo sapiens |
Nucleotide Sequence
>SYDE1
atggccgagccgctactcaggaaaaccttctcccgcctgcggggccgggagaaacttccc
cggaaaaagtcggacgccaaggagcgcggccacccagcccagcgcccagagcccagccct
ccagagccagagccccaggctcccgaagggtcccaggccggagcagaggggccctccagc
cccgaggcatcaaggagccctgcacggggagcctacctgcaaagcctggagcccagtagc
cgccgatgggtgctgggtggggccaagccagctgaggacacctctttagggcctggggta
cctggcactggggagcccgccggcgagatctggtacaaccccatccctgaggaagacccc
agacctccagcacctgagcccccggggccacagcctggctcagctgagtcagagggcctg
gccccccaaggtgcagcccccgccagccccccaaccaaagcctcccgcaccaagtccccg
ggccccgccaggcgcctctccataaagatgaagaagctgccggaactgcggcgccgcctg
agcctgcgaggcccccgggctggcagggagcgcgagagggctgcccctgcgggctccgtc
atcagccgctaccacctggacagcagcgtggggggccccgggccggcagcagggcctggg
ggcacccggagcccgagggccggttacctcagcgacggggactcaccggagcgcccagct
gggcccccatcacccacctccttccggccctacgaggtgggtcccgcagcccgggcaccc
ccggccgcactctggggccgcctcagcctgcacctgtacggtctcggggggctgcggcca
gcgccgggggccacccccagggacctctgctgcctactgcaagtggatggggaggccagg
gcccgaacagggccactgcgaggggggccggacttcctgcggctggaccacaccttccac
ctggagctggaggccgccaggctcctgcgcgccctggtgcttgcgtgggaccctggcgtg
agaaggcaccggccctgtgcccagggcaccgtgctgctgcccacggtcttccgagggtgc
caggcccaacagctggccgtgcgcctggagcctcaggggctgctgtatgccaagctgacc
ctgtcggagcagcaggaagcccctgccacagctgagccccgcgtctttgggctgcccctg
ccactgctggtggagcgggagcggccccccggccaggtgcccctcatcatccagaagtgc
gttgggcagatcgagcgccgagggctgcgggtagtgggactgtaccgtctttgtggctca
gcggcagtgaagaaagagcttcgggatgcctttgagcgggacagtgcagcggtctgccta
tctgaggacctgtaccccgatatcaatgtcatcactggcatcctcaaggattatcttcga
gagttgcccaccccactcatcacccaacccctgtataaggtggtactggaggccatggcc
cgggaccccccaaacagagttccccccaccactgagggcacccgagggctcctcagctgc
ctgccagatgtggaaagggccacgctgacgcttctcctggaccacctgcgcctcgtctcc
tccttccatgcctacaaccgcatgaccccacagaacttggccgtgtgcttcgggcctgtg
ctgctgccggcacgccaggcgcccacaaggcctcgtgcccgcagctccggcccaggcctt
gccagtgcagtggacttcaagcaccacatcgaggtgctgcactacctgctgcagtcttgg
ccagatccccgcctgccccgacaatctccagatgtcgcgccttacttgcgacccaaacga
cagccacctctgcacctgccgctggcagaccccgaagtggtgactcggccccgcggtcga
ggaggccccgaaagccccccgagcaaccgctacgccggcgactggagcgtttgcgggcgg
gacttcctgccttgtgggcgggatttcctgtccgggccagactacgaccacgtgacgggc
agtgacagcgaggacgaggacgaggaggtcggcgagccgagggtcaccggtgacttcgaa
gacgacttcgatgcgcccttcaacccgcacctgaatctcaaagacttcgacgccctcatc
ctggatctggagagagagctctccaagcaaatcaacgtgtgcctcttaatta
Protein Sequence
>SYDE1
MAEPLLRKTFSRLRGREKLPRKKSDAKERGHPAQRPEPSPPEPEPQAPEGSQAGAEGPSS
PEASRSPARGAYLQSLEPSSRRWVLGGAKPAEDTSLGPGVPGTGEPAGEIWYNPIPEEDP
RPPAPEPPGPQPGSAESEGLAPQGAAPASPPTKASRTKSPGPARRLSIKMKKLPELRRRL
SLRGPRAGRERERAAPAGSVISRYHLDSSVGGPGPAAGPGGTRSPRAGYLSDGDSPERPA
GPPSPTSFRPYEVGPAARAPPAALWGRLSLHLYGLGGLRPAPGATPRDLCCLLQVDGEAR
ARTGPLRGGPDFLRLDHTFHLELEAARLLRALVLAWDPGVRRHRPCAQGTVLLPTVFRGC
QAQQLAVRLEPQGLLYAKLTLSEQQEAPATAEPRVFGLPLPLLVERERPPGQVPLIIQKC
VGQIERRGLRVVGLYRLCGSAAVKKELRDAFERDSAAVCLSEDLYPDINVITGILKDYLR
ELPTPLITQPLYKVVLEAMARDPPNRVPPTTEGTRGLLSCLPDVERATLTLLLDHLRLVS
SFHAYNRMTPQNLAVCFGPVLLPARQAPTRPRARSSGPGLASAVDFKHHIEVLHYLLQSW
PDPRLPRQSPDVAPYLRPKRQPPLHLPLADPEVVTRPRGRGGPESPPSNRYAGDWSVCGR
DFLPCGRDFLSGPDYDHVTGSDSEDEDEEVGEPRVTGDFEDDFDAPFNPHLNLKDFDALI
LDLERELSKQINVCLLI